
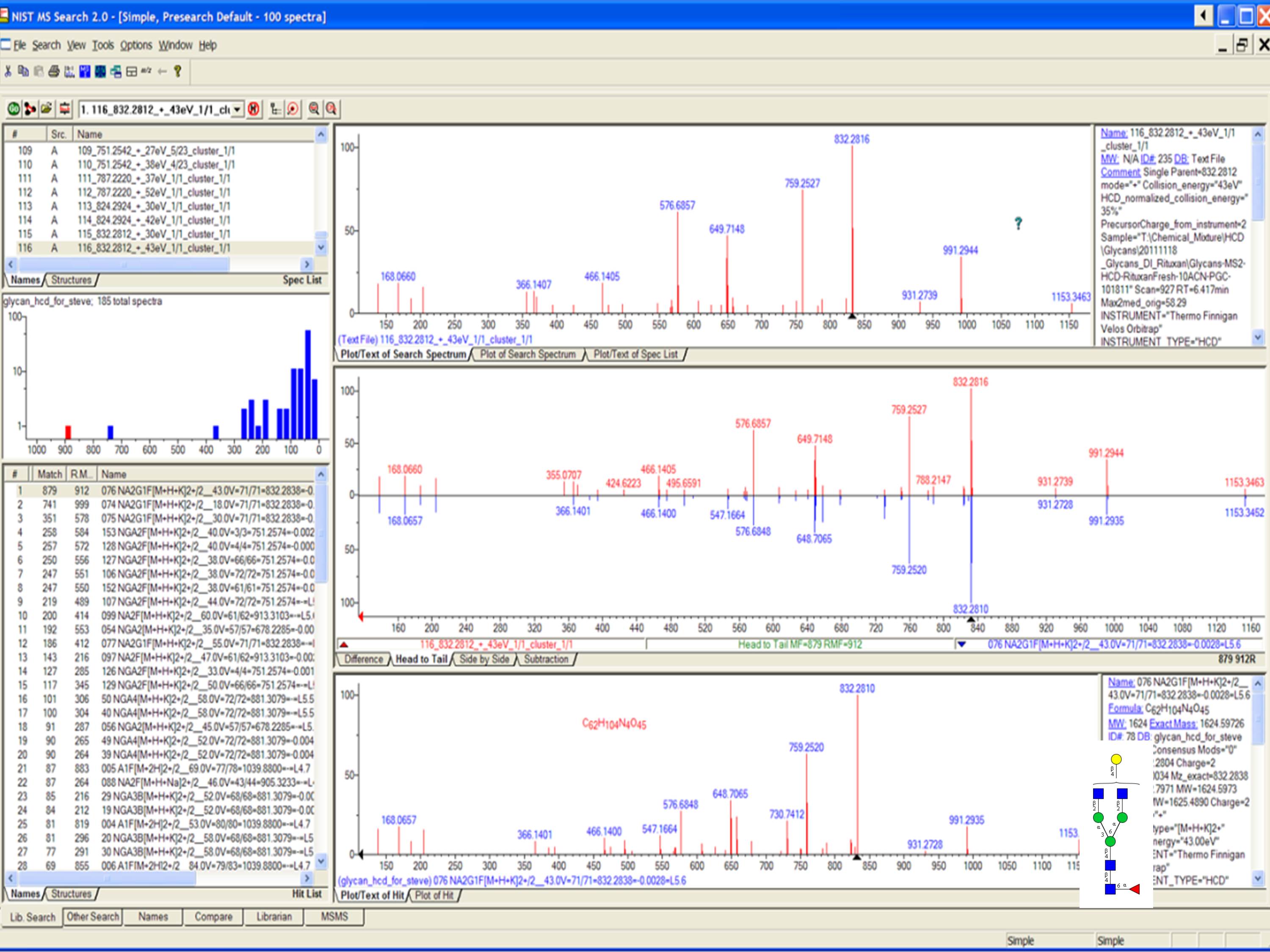

In a typical top-down analysis, more proteoforms are observed in the MS1 spectra than identified by MS2 Proteoform Suite increases the number of proteoform identifications in top-down analyses by identifying proteoforms by accurate intact mass.
#MMASS FRAGMENTATION SOFTWARE SOFTWARE#
Proteoform Suite is a software program that performs intact-mass analysis and quantification of proteoforms in mass spectrometry MS1-only data. MzLib: A library for mass spectrometry projects. We are constantly adding new features to the software, and release new versions of MetaMorpheus every few weeks. We have a community of over 200 users, which grows every day. We also focus on providing this tool to the proteomics community as a free, open-source, easy-to-use tool. Recently, we have added the ability to identify proteoforms, chemically crosslinked peptides, and isotopically-labeled peptides. MetaMorpheus was specifically designed to identify the “dark matter” of proteomics where most search programs attempt to identify mostly unmodified peptides, we provide the capability of identifying peptides with a wide range of post-translational modifications. We use computer programming as a tool to solve these problems, much like traditional chemical biology uses organic chemistry to probe biological systems. Often, the motivation for adding a feature to MetaMorpheus is the need to analyze new types of data that existing software simply cannot handle. Currently, five graduate students and several undergraduates work on improving MetaMorpheus, making it one of our most collaborative and dynamic projects. It is capable of searching both bottom-up (peptide) and top-down (intact proteoform) data. MetaMorpheus is a software tool that identifies peptides, proteins, and proteoforms from mass spectrometry fragmentation (MS/MS) data. The primary goal of the software is to provide a fast, high-quality, quantitative platform to quantify any search software’s identifications. As the name implies, it is orders of magnitude faster than existing quantification software. This step is performed after these analytes have been identified by a search program, such as MetaMorpheus. User interface making it easy to interact with the program.FlashLFQ is a program that quantifies peptides, proteins, and proteoforms in mass spectrometry data. Abacus is compatible with the widely used Trans-Proteomic Pipeline suite of tools and comes with a graphical It can also output the spectral count data at the gene level, thus simplifying the integration and comparison between

It aggregates dataįrom multiple experiments, adjusts spectral counts to accurately account for peptides shared across multiple proteins, and performs common Curr Protoc Bioinformatics 2012, Chapter 8, Unit8.15.Ībacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysisĪBACUS is a computational tool for extracting label-free quantitative information (spectral counts) from MS/MS data sets. Analyzing Protein-Protein Interactions from Affinity Purification-Mass Spectrometry Data with SAINT. SAINTexpress is a recently developed fast version ofĬhoi, H. The method was initially developed for label-free spectralĬount data, but was later extended to MS1 intensity-based quantitative data (SAINT-MS1). Incorporates various data normalization steps and is also capable of utilizing the quantittaive information from negative control purificationsįor improving specificity in small-to-intermediate scale experiments (SAINT v. SAINT: probabilistic scoring of affinity purification–mass spectrometry dataĬomputational models and software for assigning confidence scores to protein-protein interactions in label-free quantitative AP-MS datasets.įor each observed interaction with associated label-fee quantification, SAINT calculates the probability of true interaction.


 0 kommentar(er)
0 kommentar(er)
